Introduction
Huntington's disease (HD) is an autosomal dominant neurodegenerative disorder caused by a polyglutamine repeat expansion within the huntingtin protein. HD is characterized by problems with movement, cognition and behavioral functioning, and there is currently no effective treatment. Although multiple pathologic mechanisms have been proposed, the exact mechanism by which mutant huntingtin causes neuronal dysfunction is not known. Recent studies demonstrating altered messenger RNA expression point to transcriptional dysregulation as a central mechanism. The control of eukaryotic gene expression depends on the modification of histone proteins associated with specific genes, with histone acetylation playing a crucial role. Studies in numerous HD models have shown that mutant huntingtin alters histone acetyltransferase activity, and indicate that aberrant activity of this enzyme might be an underlying mechanism of transcriptional dysregulation in HD. Furthermore, recent studies have shown a therapeutic role for histone deacetylase inhibitors in a number of HD models. In this review, we summarize the current state of knowledge regarding the status of histones in HD. In addition, we discuss how these histone modifications not only lead to pathogenesis, but might also provide a novel therapeutic strategy for treating this devastating disease.
Huntington's disease (HD) is an autosomal dominant neurological disease that afflicts around one in 10,000 people. HD onset typically occurs in young adult life, and the disease is invariably fatal. Patients initially develop small choreic movements that later intensify in amplitude and frequency, causing an impairment of daily activities. Although the movement disorder is typically chorea, other types of movement disorder, such as DYSTONIA, rigidity, MYOCLONUS and ATHETOSIS, can also be seen. Psychiatric disturbances, the most common of which are apathy, depression, obsessivecompulsive disorder, psychotic disorders, substance abuse and paranoia, are often the most disabling symptoms for patients with HD. Cognitive disorders, on the other hand, are among the most difficult symptoms to treat. They appear later in the course of the disease, manifesting as dementia and difficulties in executive functioning. Weight loss is also a common feature in HD, implying the existence of a generalized metabolic defect. To date, there are no effective treatments to cure the disease or to slow its progression.
Huntingtin (Htt), the protein that is defective in HD, is expressed throughout the body, yet HD principally affects the brain. Although HD affects a number of regions, including the cortex, thalamus and subthalamic nucleus, the striatum is the most severely affected region. Within the striatum, the medium spiny projection neurons are preferentially targeted by the disease, whereas interneurons are relatively spared. The preferential loss of medium spiny neurons and sparing of interneuron populations indicates that neurodegeneration in HD is cell-type-specific. The factors that render striatal projection neurons more susceptible to damage are, however, currently unknown.
Transcriptional dysregulation has been proposed to have an important role in the pathology of HD, but the mechanisms that cause disruption of gene expression remain unknown. The control of eukaryotic gene expression depends on the modification of histone proteins associated with specific genes, with histone acetylation playing a crucial role. Acetylation of histones at specific residues increases gene transcription; conversely, histone deacetylation represses transcription. Recent studies in numerous HD models have demonstrated a potential therapeutic role for histone deacetylase (HDAC) inhibitors in the treatment of polyglutamine diseases. In this review, we summarize these studies, and discuss how histone modifications might provide a novel therapeutic approach for treating HD.
The Huntington's Disease Mutation
HD is caused by a mutation in the IT15 gene on the short arm of chromosome 4. This gene, which was subsequently renamed HD, consists of 67 exons that encode Htt, a 350-kD protein of 3,144 amino acids. The mutation is an expansion of the cytosineadenineguanine (CAG) trinucleotide repeat in exon 1, which codes for a polyglutamine moiety in the Htt protein. Normal individuals have CAG repeat lengths of 734. The CAG repeat is expanded and unstable in HD patients, with repeat length inversely correlating with age of disease onset. Repeat lengths of more than 40 glutamines produce HD, and repeats of over 70 glutamines invariably cause juvenile onset.
When the HD mutation was eventually identified as a CAG trinucleotide repeat expansion, HD joined a novel class of neurodegenerative disease, the polyglutamine diseases. In all of these disorders, expansion of the CAG repeat occurs within the coding region of the gene, so the CAG repeat is translated into a polyglutamine stretch. Polyglutamine diseases are all autosomal dominant or sex-linked dominant diseases, usually with adult onset, and are characterized by progressive neurodegeneration of selected neuronal populations. In both humans and transgenic mouse models, these disorders are characterized pathologically by polyglutamine protein-containing intracellular inclusions.
The discovery of the Huntington's disease gene in 1993 resulted in a direct genetic test to make or confirm a diagnosis of Huntington's disease in an individual who is exhibiting Huntington's disease-like symptoms. Using a blood sample, the genetic test analyzes DNA for the Huntington's disease mutation by counting the number of repeats in the Huntington's disease gene region. Individuals who do not have Huntington's disease usually have 28 or fewer CAG repeats. Individuals with Huntington's disease usually have 40 or more repeats. A small percentage of individuals, however, have a number of repeats that fall within a borderline region
Pathogenic Mechanisms
The genetic mutation that causes HD was discovered in 1993, but the mechanism by which mutant Htt causes neuronal dysfunction is still not known. Several theories have been proposed to explain the mechanism of HD pathogenesis. Excitotoxicity has been proposed as a pathogenic mechanism, on the basis of the observation that infusion of glutamate-receptor agonists into the brain leads to neuronal death and a phenotype similar to HD. Mitochondrial dysfunction is another leading hypothesis, as numerous deficits in mitochondrial metabolism have been reported in the brains of patients with HD at postmortem examination. Caspases, a family of proteases that are involved in apoptotic cell death, show increased activity in HD, and are also thought to participate in HD pathogenesis. Furthermore, caspases can generate amino-terminal (N-terminal) fragments of Htt that can enter the nucleus and form nuclear inclusions. Beside caspases, other types of proteolytic cleavage have been identified, involving calpain, AUTOPHAGY, proteasomes or an aspartyl protease. Nuclear inclusions formed by mutant Htt sequester proteins such as transcription factors, heat-shock proteins and proteasome subunits, thereby preventing these proteins from reaching their necessary point of action.
In addition to the formation of nuclear inclusions, mutant Htt can also form toxic β-sheet-rich aggregates through the misfolding of the expanded polyglutamine stretch. This aggregation process appears to be associated with pathogenesis in HD. Polyglutamine expansion interferes with proteasome function, and polyglutamine proteins might disrupt the global balance of protein-folding control.
Autophagy has emerged as a mechanism of HD pathogenesis on the basis that mutant Htt accumulation activates the endosomallysosomal system and contributes to Htt proteolysis and autophagic cell death. The role of autophagy is unclear, however. Inhibition of mTOR (mammalian target of rapamycin; also known as FRAP1) activates autophagy and attenuates Htt-induced toxicity, indicating that autophagy might in fact be helpful as a clearance mechanism.
Transcriptional Dysregulation in Huntington's Disease
Recent studies have provided strong evidence that transcriptional dysregulation is an important underlying mechanism in HD pathogenesis. Transcriptional dysregulation is an early event in HD pathology, and is observed across multiple HD models. The polyglutamine repeats in the N-terminal region of Htt protein gives it structural similarities to known transcription factors, and repeat expansion leads to aberrant cleavage by caspases. The cleaved fragments gain access to the nucleus and form nuclear aggregates that might disrupt transcription. Mutant Htt interacts with numerous transcription factors including CREB-binding protein (CBP), TATA-binding protein (TBP), p53 and Sp1, raising the possibility that Htt nuclear aggregates cause transcriptional dysregulation by sequestering transcription factors.
Humans with HD and transgenic mouse models of HD show downregulation of specific genes at the level of messenger RNA (mRNA) expression. The R6 mouse lines in particular have provided evidence for transcriptional dysregulation. These mice were created by inserting exon 1 of the human HD gene containing 150 polyglutamine repeats, under the control of the human HD promoter, into the mouse genome. Alterations of neurotransmitter receptor protein levels, as well as of mRNA levels, have been reported in the R6/2 mouse line. Both of these changes occurred before the onset of abnormal symptoms. This finding is in agreement with observations in patients in the early stages of HD, in that the pathologic changes appear to precede the appearance of disease symptoms. PET studies of gene-positive but clinically asymptomatic patients in the early stages of HD demonstrate that expression of dopamine D1 and D2 receptors is decreased before the onset of symptoms. Therefore, neuronal dysfunction predates the appearance of neurological symptomatology in HD. In very early-grade HD cases (grade 0), there are profound reductions in the expression levels of cannabinoid CB1, dopamine D2 and adenosine A2a receptors.
Widespread screening of mRNA levels, using DNA microarrays, in the brains of R6/2 mice confirmed the finding that specific genes are downregulated in this HD mouse model. The transcripts affected are important for neuronal function, and they include genes involved in neurotransmitter signaling, calcium metabolism and transcription. Decreases in mRNA levels were more common than increases, and the set of genes that were decreased at 12 weeks of age was larger than the set of genes that were affected at 6 weeks, indicating a progressive effect.
Gene expression studies have also been carried out in other HD mouse models. The HD-N171-82Q transgenic mouse model expresses a COMPLEMENTARY DNA that encodes a 171-amino-acid N-terminal fragment of Htt exon 1 containing 82 CAG repeats. The changes in gene expression in HD-N171-82Q mice are similar to those observed in R6/2 mice. The mRNA changes observed in the R6/2 and HD-N171-82Q mice are of significance, given that they recapitulate the gene expression changes that occur in the human HD brain. Expression profiling of human HD cases shows that gene changes are most pronounced in the striatum, with the motor cortex affected to a lesser degree, and the cerebellum showing the fewest gene changes.
R6/2 and HD-N171-82Q mice are fragment models; that is, they express truncated portions of Htt. Transgenic mouse models expressing full-length versions of Htt, such as the YAC72 mice, demonstrate fewer gene changes, indicating that increasing the length of Htt reduces the severity of polyglutamine-induced gene changes.
Mutant Htt interacts with numerous transcription factors, providing a mechanism through which this protein can interfere with normal transcriptional activity. For example, soluble mutant exon 1 Htt protein has been shown to interact with the polyQ-containing domain of CBP. Recently, Zhai and colleagues have shown that mutant versions of N-terminal Htt can dissociate components of the transcriptional complex on gene promoters. Transcriptional dysregulation has also been proposed as a disease mechanism in other polyglutamine diseases. In fact, the androgen receptor, which is mutated in spinobulbar muscular atrophy or Kennedy's disease, and the TATA box binding protein (TBP), which is mutated in spinocerebellar ataxia type 17 (SCA-17), are themselves transcription factors. Ataxin-7, the protein mutated in SCA-7, is a component of the mammalian STAGA (SPT3-TAF9-ADA-GCN5 acetyltransferase) transcription coactivator complex, and it interacts directly with the GCN5 histone acetyltransferase. Taken together, these results indicate that transcriptional dysregulation is likely to be an important mechanism in the pathogenesis not only of HD, but also of other polyglutamine disorders.
If downregulation of the expression of key molecules at the mRNA level underlies the neuronal dysfunction in HD, correction of these transcriptional abnormalities has great potential as a novel therapeutic approach. To harness this potential, however, it will be necessary to understand the mechanisms that cause selective downregulation of target genes.
Histone Modifications
Regulation of gene expression is accomplished through the action of transcription factors that alter chromatin structure through the recruitment of histone-modifying enzymes. The N-terminal tails of the core histones (H2A, H2B, H3 and H4) are strongly basic, and contain specific amino-acid residues that are sites for several post-translational modifications, including acetylation, methylation, phosphorylation, ubiquitination and SUMOYLATION . These modifications can affect processes such as transcription, mitosis and chromosome stability. In general, acetylation of lysine residues corresponds to transcriptionally active chromatin, whereas methylation of lysine and arginine residues leads to transcriptional repression. For example, acetylation of lysine 9 and 14 on histone H3 (AcK9K14H3) correlates with active chromatin and leads to transcription, whereas methylation of lysine 9 on histone H3 (MeK9H3) is a marker of silenced genes. Phosphorylation of serines also correlates with transcriptional activation, because it precedes acetylation at lysine residues. Many combinations of covalent modifications are possible on the core histones, and these precise patterns of histone modification seem to be a key factor in turning specific genes on or off. This system of gene control has been termed the 'histone code' .
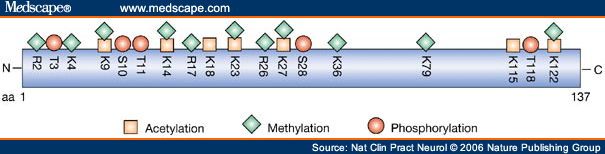
Known modifications of human histone H3. Acetylation and methylation at lysine residues are mutually exclusive, and lysine residues can be monomethylated, dimethylated or trimethylated. aa = amino acid; K = lysine; R = arginine; S = serine; T = threonine.
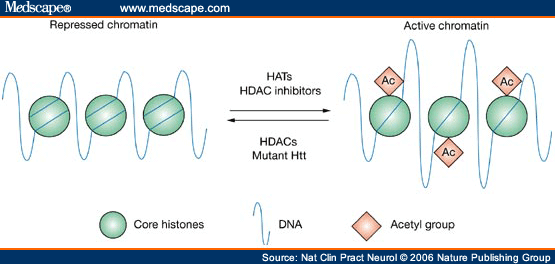
Histone acetylation and deacetylation are modulated by the interplay between histone acetyltransferases (HATs) and HDACs, which work in concert to modify chromatin structure and regulate transcription. In simple terms, HAT activity leads to increases in gene transcription by creating a more open conformation of chromatin, whereas HDACs remove acetyl groups, leading to gene repression through condensation of chromatin. In reality, however, the relationship between histone acetylation and gene transcription is likely to be more complex. An emerging view is that histone acetylation not only governs the relaxation of chromatin, but also regulates the recruitment of specific transcriptional regulatory complexes. Therefore, rather than directly correlating with activation of gene expression, histone acetylation might render chromatin 'transcription-competent'. Abnormalities of histone acetylation have been associated with a number of human cancers; for example, in leukemia and non-Hodgkin's lymphoma, mutations activate and target HDACs. In human gastrointestinal cancers, histone acetylation is globally reduced.
Acetylation of core histone proteins through the activity of histone acetyl transferase proteins or treatment with histone deacetylase inhibitors leads to a more open conformation of chromatin, and therefore a transcriptionally active state. Removal of acetyl groups by histone deacetylases leads to the formation of repressed chromatin, which corresponds with transcriptional repression. Similarly, mutant huntingtin (Htt) expression is believed to drive the formation of repressed chromatin by decreasing acetylation of histones. HAT = histone acetyl transferase; HDAC = histone deacetylase.
Gardian and colleagues administered the HDAC inhibitor phenylbutyrate after the onset of symptoms in another transgenic mouse model, HD-N171-82Q. Phenylbutyrate administered to HD-N171-82Q mice at 75 days of age increased survival, and decreased striatal atrophy and ventricular enlargement. There was no effect on motor performance, weight loss or Htt aggregate formation. Histone H3 and H4 acetylation was increased in the striatum following phenylbutyrate treatment, and there was a decrease in methylation of histone H3. Microarray analysis of the phenylbutyrate-treated HD-N171-82Q striatum demonstrated that some genes were upregulated and others were downregulated. Although phenylbutyrate did not improve the expression of mutant-Htt-downregulated genes, it is promising that transgenic mice showed an overall improvement in their condition, given that treatment began after the onset of symptoms.
.
The therapeutic mechanism of HDAC inhibitors is not clear. The simplest mechanism that can be envisaged is one in which administration of HDAC inhibitors corrects the downregulation of specific genes caused by mutant Htt. Limited studies of HDAC inhibitor treatment of transgenic mice, however, indicate that this is not the case. An alternative possibility is that the benefits of HDAC inhibitors derive from global increased gene expression. If this is the case, HDAC inhibitors might be beneficial in other neurodegenerative conditions beyond HD. Indeed, HDAC inhibitors have also shown promise in transgenic mouse models of amyotrophic lateral sclerosis, schizophrenia and ischemia.
There are three classes of HDACs: class I and class II HDACs have been classified on the basis of sequence similarities, whereas class III HDACs are a group of NAD-dependent deacetylase enzymes related to the yeast Sir2 protein. The role of each individual HDAC in regulating gene expression is not known. Some HDACs deacetylate other proteins in addition to histones. In addition, there are tissue-specific and cell-type-specific differences in HDAC expression, localization and targets. Therefore, an individual HDAC inhibitor might have different effects in different biological systems. HDAC inhibitors are toxic and can induce cell cycle arrest by increasing the transcription of p21 and p53, so it will be important to target individual HDACs to minimize cytotoxicity. Other side effects are associated with HDAC inhibitor therapy; for example, prolonged TSA therapy enhances chromosomal instability, leading to defective centromeres and abnormal chromosomal segregation.In addition, HDAC inhibitors can promote tumor development in some cases. Therefore, it will be vital to determine the toxicity of these compounds.
In August 2008 the U.S. Food and Drug Administration approved tetrabenazine to treat Huntington's chorea, making it the first drug approved for use in the United States to treat the disease. Antipsychotic drugs, such as haloperidol, or other drugs, such as clonazepam, may help to alleviate choreic movements and may also be used to help control hallucinations, delusions, and violent outbursts. Antipsychotic drugs, however, are not prescribed for another form of muscle contraction associated with Huntington's disease, called dystonia, and may in fact worsen the condition, causing stiffness and rigidity. These medications may also have severe side effects, including sedation, and for that reason should be used in the lowest possible doses.
For depression, physicians may prescribe fluoxetine, sertraline, nortriptyline, or other compounds. Tranquilizers can help control anxiety and lithium may be prescribed to combat pathological excitement and severe mood swings. Medications may also be needed to treat the severe obsessive-compulsive rituals of some individuals with Huntington's disease.
Most drugs used to treat the symptoms of Huntington's disease have side effects such as fatigue, restlessness, or hyperexcitability. Sometimes it may be difficult to tell if a particular symptom, such as apathy or incontinence, is a sign of the disease or a reaction to medication.
Conclusions
Currently, HD remains a fatal untreatable disease. Recent advances in understanding the underlying pathologic mechanisms, however, have provided hints to the development of effective therapies. Specifically, recent evidence points strongly to transcriptional dysregulation as an important mechanism. The alteration of gene expression is likely to occur as a consequence of abnormal histone modifications, including hypoacetylation. The use of HDAC inhibitors and other therapies that target gene transcription is an exciting development in the field of HD therapeutics. There are strong indications that HDAC inhibitors might be of therapeutic benefit in HD, but their precise mechanism of action has yet to be determined.