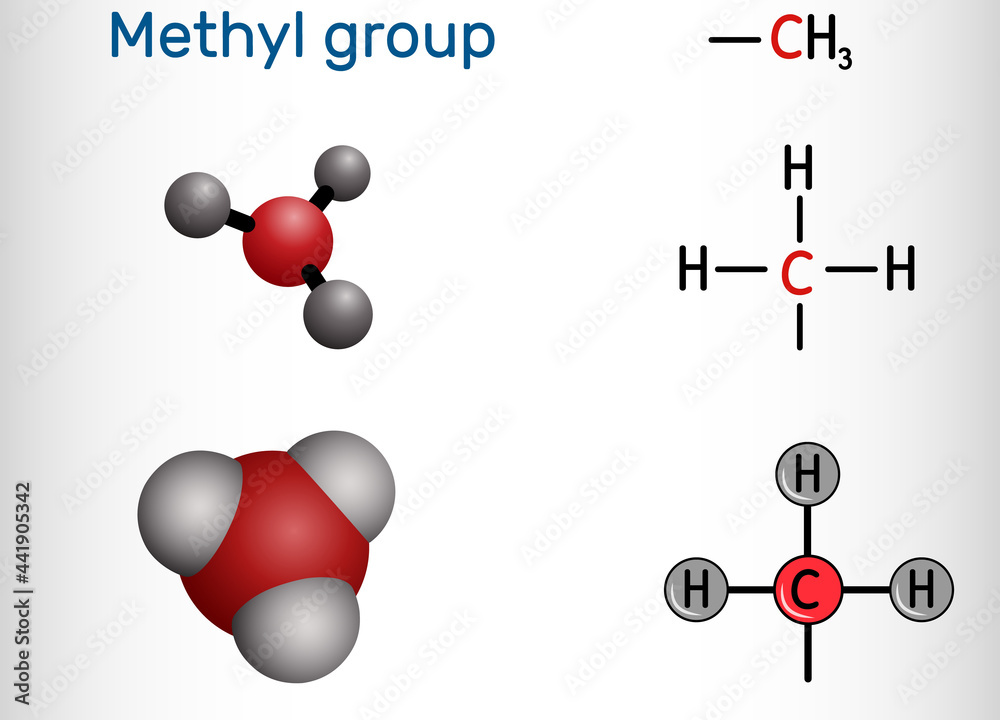
Methyl Group
Methylation targets are:
- DNA
- RNA
- Proteins
- Lipids
- Hormones and neurotransmitters (COMT, HIMT)
- Telomere
Methyl Donor

Methyl Transferases
DNA
DNA methylation is a crucial epigenetic modification of the genome that is involved in regulating many cellular process. These include, embryonic development, transcription, chromatine structure, X chromosome inactivation, genomic imprinting and chromosome stability. (Robertson 2005 Nat. Rev.)

Methylation contributing to epigenetic inheritance can occur either through DNA methylation or protein methylation
DNA methylation in vertebrates typically occurs at CpG sites (cytosine-phosphate-guanine sites; that is, where a cytosine is directly followed by a guanine in the DNA sequence); this methylation results in the conversion of the cytosine to 5-methylcytosine. The formation of Me-CpG is catalyzed by the enzyme DNA methyltransferase. The bulk of mammalian DNA has about 40% of CpG sites methylated but there are certain areas, know as CpG islands which are GC rich (made up of about 65% CG residues) where none are methylated. These are associated with the promoters of 56% of mammalian genes, including all ubiquitously expressed genes. 1-2% of the human genome are CpG clusters and there is an inverse relationship between CpG methylation and transcriptional activity.

DNA methylation
Azacytidine-induced tumorigenesis of CHEF/18 cells: Correlated DNA methylation and chromosome changes, 1983
RNA
Proteins
Protein methylation is one type of post-translational modification.
Protein methylation typically takes place on arginine or lysine amino acid residues in the protein sequence.
Identification, Quantification, and System Analysis of Protein N-ε Lysine Methylation in Anucleate Blood Platelets, 2019
charcot+marie+tooth+and+proteins+methylation
Protein Aminoacids Percentage (Width 700 px)


Proteins transferring CH3 to Lysine are older than those transferring to Arginine
DNA methylation is a crucial epigenetic modification of the genome that is involved in regulating many cellular process. These include, embryonic development, transcription, chromatine structure, X chromosome inactivation, genomic imprinting and chromosome stability. (Robertson 2005 Nat. Rev.)

Methylation contributing to epigenetic inheritance can occur either through DNA methylation or protein methylation
DNA methylation in vertebrates typically occurs at CpG sites (cytosine-phosphate-guanine sites; that is, where a cytosine is directly followed by a guanine in the DNA sequence); this methylation results in the conversion of the cytosine to 5-methylcytosine. The formation of Me-CpG is catalyzed by the enzyme DNA methyltransferase. The bulk of mammalian DNA has about 40% of CpG sites methylated but there are certain areas, know as CpG islands which are GC rich (made up of about 65% CG residues) where none are methylated. These are associated with the promoters of 56% of mammalian genes, including all ubiquitously expressed genes. 1-2% of the human genome are CpG clusters and there is an inverse relationship between CpG methylation and transcriptional activity.

DNA methylation
Protein methylation typically takes place on arginine or lysine amino acid residues in the protein sequence. Arginine can be methylated once (monomethylated arginine) or twice, with either both methyl groups on one terminal nitrogen (asymmetric dimethylated arginine) or one on both nitrogens (symmetric dimethylated arginine) by peptidylarginine methyltransferases (PRMTs). Lysine can be methylated once, twice or three times by lysine methyltransferases. Protein methylation has been most well studied in the histones. The transfer of methyl groups from S-adenosyl methionine to histones is catalyzed by enzymes known as histone methyltransferases. Histones which are methylated on certain residues can act epigenetically to repress or activate gene expression. Protein methylation is one type of post-translational modification.
Asimmetric dimethyl arginine
“Telomere”: https://flipper.diff.org/app/pathways/Telomere methylation
Lipids
Hormones and neurotransmitters