Background
Chemosensation is perhaps the most ancient yet enigmatic sense. All organisms living on Earth, from simple unicellular forms to complex multicellular creatures possess the capability to detect chemicals in the surroundings. Evolutionally, many species, from insects to mammals, have developed the olfactory system, on which they rely to locate food, mates and to avoid danger. this is due to the presence of a large number of chemoceptors in in this structure, that allows them to gather information on the surroundings. This information is carried by odorants.
The vertebrate olfactory system has a remarkable capacity to discriminate and recognize a wide variety of odorants. These, binding to the chemoceptors present on the peripheral olfactory neurons in the olfactory epithelium, begin the signal transduction inside the cell. This signal reaches the olfactory bulb of the brain then, whose task is to interpret the stimulus. What is not yet fully understood are the mechanisms behind an enhanced individual sensitivity to certain odorants.
 Human olfactory system. 1. Olfactory bulb 2. Mitral cells 3. Bone 4. Nasal epithelium 5. Glomerulus 6. Olfactory receptor neurons.
Human olfactory system. 1. Olfactory bulb 2. Mitral cells 3. Bone 4. Nasal epithelium 5. Glomerulus 6. Olfactory receptor neurons.
Introduction
Over the past two decades special attention has been directed at the human olfactory system, as its dysfunctions deeply impact the individual's life. For example, olfactory loss - anosmia - has been proven to be a very common symptom of chronic rhinosinusitis (CRS). Anosmia can be both temporary, caused by nasal congestion or obstruction as in the flu or some types of rhinitis, or permanent, mainly due to brain injuries or death of olfactory receptor neurons. In this last case, the loss of smell may lead to harmful conditions such as depression and such hazardous behaviours as involuntarily eating spoiled food, or not recognizing dangerous liquids and gases.
While the causes of anosmia have been widely studied due to their medical importance, not the same amount of attention has been paid to the other side of the coin: oxyosmia, also known as hyperosmia, an individual increased olfactory acuity to a single odorant or to a group of them.
Hyperosmia is a complex trait that has not yet been fully understood, mainly because there seem to be no complete studies on the matter. Many hypotheses have arisen that may explain such an interesting phenomenon: generally speaking, hyperosmia is supposed to be due to individual variations regarding either the olfactory receptor neurons (ORN), and specifically the olfactory receptors (OR) and the signalling pathway, or the olfactory bulb, where the signals are interpreted. Of course, such variations may affect both structures. Evidence shows that hyperosmia could be mainly due to polymorphisms inside the aminoacid structure of the ORs, and this hypothesis has the virtue of simplicity.
Genetic Causes
The functional characterization of odorant receptors conducted over the past decade has led to the belief that the molecular base of odor discrimination lies in the interaction of an odorant with an OR. A single OR can recognize multiple odorants sharing similar structures, and different odorants are recognized by multiple ORs with various degrees of affinity, but most important, different odorants are recognized by distinct combinations of ORs. That means that every single odorant has a peculiar receptor code. Moreover, these codes are partially overlapping. In this way, a slight change in the structure of the odorant can produce a complete change in the perceived smell. In the olfactory epithelium each ORN expresses a single allele of a single OR gene, and the OR genes form a multigene family consisting of over 850 loci in humans, many of which are considered non-functional due to frame interrupting mutations. The reason behind this huge amount of pseudogenes is to be sought in a less dependency of primates on olfactory clues. It is mainly an evolutionary process, and the interesting part of it is that it has been demonstrated that OR genes mostly segregate between intact and pseudogene forms: each individual is characterized by a particular combination of segregating pseudogenes. This consitutes part of the great genotypic diversity in the population. For this amount of genetic material involved, it is widely accepted that allelic variants of OR genes may determine different functional characteristics of ORs and therefore result in different odorant sensitivity phenotypes in the human population (Olender et al. See below).
A fairly recent study (Menashe et al. Genetic elucidation of human hyperosmia to isovaleric acid (IVA), 2007.) investigates the genetic basis for odorant specific variations in different individuals using OR segregating pseudogenes, which have both functional and nonfunctional alleles in the population. The aim of this study was to evaluate the effect of single nucleotide polymorphisms (SNPs) of 43 OR segregating loci in terms of odorant-specific olfactory threshold variations. A statistically significant association between the presence of a nonsense SNP in the coding region of OR gene OR11H7P and sensitivity for the odorant IVA was found. Individuals with specific hyperosmia to this odorant are more likely to be heterozygous or homozygous intact for this gene, whereas homozygous disrupted could not detect IVA at low concentration. This association was later verified in vitro on Xenopus (frog) oocytes, where the active allele was expressed first. Its response was then tested at various IVA concentrations (ranging from 30 microM to 10 milliM) and later compared to that of the inactive allele, showing that there was a significant difference (p<0.05) in the response between the two alleles. In this study it was also shown that a higher sensitivity for a specific odorant predicts a tendency of the same individual to have a lower threshold (higher sensitivity) for other odorants. This may be related to an increased affinity of the OR in question for other odorants as well. On the other hand, an appealing possibility is the contribution of genetic polymorphisms involving the signal transduction genes. The results presented in this study show an initial link between the incredibly high genetic variability in OR genes and individual variations in olfaction.
 OR genes on chromosome 14. Red: pseudogenes. Yellow: Segregating Pseudogenes (SPGs). Green: Intact genes. Marked with asterisks are those SPGs showing strongest association with IVA sensitivity. Image taken from Menashe et al. study.
OR genes on chromosome 14. Red: pseudogenes. Yellow: Segregating Pseudogenes (SPGs). Green: Intact genes. Marked with asterisks are those SPGs showing strongest association with IVA sensitivity. Image taken from Menashe et al. study.
Further insight into this topic has been provided by another study, where genetic variations in 413 intact OR loci were considered, for which at leas one individual had an open reading frame (ORF) (Olender et al. Personal receptor repertoires: olfaction as a model, 2012). The aims of this study were to provide a valuable inventory of the human olfactory receptors coding regions on which to base further studies on the matter. In the first step of the study, genomic variations data in OR genes were obtained from 1000 Genome Project and other sources and were used to highlight the numerous allelic variants in the OR coding regions: for the 413 intact OR loci, about 5950 polymorphic variations were found. From this data, 4069 putative haplotypic OR alleles were identified, with an average of ca. 10 variations per locus. These variants represent distinct funcional proteins with possibly different affinity or specificity for odorants. Each individual is thought to present up to ca. 600 haplotypes in the 413 loci analyzed. Interestingly these haplotypes have different distributions inside the three ethnicities studied - Africans, Asians and Europeans- with the highest average of ca. 557±13 in Africans, concordant with the idea of African origin of the modern human population.
 Status diagram of the human OR repertoire. I: intact genes. P: pseudogenes. S: segregating pseudogenes. D: CNV deletion allele. R: resurrected pseudogenes (pseudogenes for which an active allele has been found in the population. Therefore they may be considered "resurrected" from their fixed pseudogene status). Image taken from Olender et al. study.
Status diagram of the human OR repertoire. I: intact genes. P: pseudogenes. S: segregating pseudogenes. D: CNV deletion allele. R: resurrected pseudogenes (pseudogenes for which an active allele has been found in the population. Therefore they may be considered "resurrected" from their fixed pseudogene status). Image taken from Olender et al. study.
Furthermore, Olender et al. were able to elaborate a status diagram of the human OR repertoire, pointing out that more then 50% of the human OR loci are considered non-functional. Interestingly, 56% ca. of the intact loci show signs of segregation between intact and non-functional alleles, making them excellent candidates for the explanation of inter-individual odorant perception. In addition to this, copy number variations (CNVs) seem to impact these huge differences in OR loci, though in a less significant way: the most likely to contribute to a detectable phenotype are deletion CNVs affecting consecutive intact ORs in a cluster, though the evolutionary meaning of this phenomenon remains unclear. This study supports the idea that every individual possesses a characteristic pattern of intact and inactive alleles, or in other words different noses for different people (Keller, Different noses for different mice and people, 2012 ): it is suggested that a randomly selected pair of individuals will share differences on average in one third of their OR loci, thus leading to the different perception we have for odorants, both in terms of strength and pleasure.
Signal Transduction
While for many aspects our knowledge regarding OR genes remains partial and needs further studies, strong evidence has proved that ORs are G protein-coupled receptors, therefore consisting of seven transmembrane domains, of which the third, the fourth and the fifth are those believed to directly interact with the odorant. The olfactory epithelium-specific G protein - composed of three subunits: α, β and γ - belongs to the G-Alpha subclass and is called G-Alpha-Olf. As of today, the signalling pathway has been widely studied and few processes remain unclear.
As odorants enter the nasal cavity, they dissolve in the mucus covering the luminal surface of the olfactory epithelium. Subsequently they bind to ORs with various degrees of affinity for their particular structure, therefore each odorant activates a particular set of ORs. The molecular features of the odorant are then transduced into electrical signals and then sent to the olfactory bulb thorugh the olfactory nerves. This transduction uses G protein-coupled second messengers.
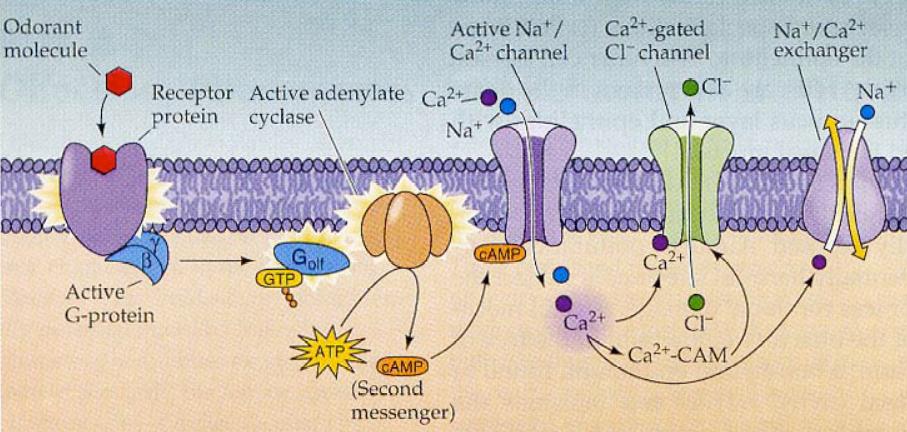
In the ORN, the GDP-bound form of G-Alpha-Olf si coupled to its OR. The binding of an odorant to the recptor activates the G-protein, which liberates GDP and binds GTP, entering its active form. The Gα-GTP subunit activates an olfactory epithelium-specific Adenylyl Cyclase, ACIII, which catalyzes the conversion of ATP to 3,5 cAMP. The increase in cAMP opens olfactory Cyclic Nucleotide-Gated channels (oCNG channels), allowing both Ca2+ and Na+ to enter the cell through the cilia. This causes a membrane depolarization, leading to an action potential transducing the signal to the olfactory bulb.
Later, Ca2+ is extruded out of the cell thorugh many ways: ClCn Channels, Na+/Ca2+ exchangers and Ca2+ ATPases. These mechanisms allow the cell to return to its original electrical neutrality and quit the signal. Another way to modulate membrane depolarization is thorugh Calmodulin (Calm), a calcium binding protein that regulates changes in intracellular Ca+2 concentrations. As soon as the Ca2+/Calm complex is formed, it binds to a particular binding site on the oCNG channel. This interaction reduces the sensitivity of the channel to cAMP and makes it close again. In addition to this, Ca2+/Calm further activates a Phosphodiesterase (PDE) that converts cAMP to 5' AMP.
The presence of other second messengers has long been debated. In particular, a few studies showed evidence of IP3 action in the olfactory system of different animals, especially in the lobster, suggesting the presence of GPCR-activated PI3-Kinase. In a recent study ( Koley et al. Phosphoinositide 3-Kinase mediated signaling in lobster olfactory receptor neurons, 2010 ) the presence and the active role of IP3 in the lobster's olfactory epithelium has been demonstrated, as well as in vitro activation of PI3K after interaction with odorants. Still, this argument need further studies, as there is no clear evidence of such a mechanism in the human olfactory system.
Conclusions
Thanks to the evidence proposed in recent studies, it is possible to demonstrate that an individual enhanced sensitivity to certain odorants may be due to genetic variations in the OR loci, which are highly polymorphic, though this unsual diversity has not yet been fully explained.
These variations reflect different aminoacid sequences in the ORs, probably mostly located in the third, fourth and fifth transmembrane domains. This results in a higher affinity for a specific odorant, granting more stable interactions between OR and molecule. The presence of these high affinity ORs in the specific receptor code of an odorant allows it to be recognized at lower concentrations, thus permitting the genesis of an action potential directed to the olfactory bulb. As a single OR binds different but structurally similar odorants, a variation in its sequence may also condition how these others are perceived.
As of today, there is no evidence that genetic variations regarding secondary messenger enzymes are related in any way to a variation in odorant detection threshold. This aspect could merit further study.
Olfaction is a complex trait which cannot be fully explained through genetics: many other factors - such as smoking, general lifestyle, ethinicity- have to be taken into account. To what degree genetic factors influence an individual's olfactory sensitivity in contrast to environmental ones has yet to be determined.