DEFINITION

DISC1 (Disrupted in schizophrenia 1) is a protein that is encoded by the DISC1 gene in humans. His weight is 93,611 Da.
DISC1 has been referred to as a 'scaffold protein' because of its ability to bind to a wide array of other proteins, including those of importance for neurodevelopment.
In coordination with a wide array of interacting partners, DISC1 has been shown to participate in the regulation of cell proliferation, differentiation, migration, neuronal axon and dendrite outgrowth, mitochondrial transport, fission and/or fusion, and cell-to-cell adhesion..
Several studies have shown that unregulated expression or altered protein structure of DISC1 may predispose individuals to the development of schizophrenia, clinical depression, bipolar disorder, and other psychiatric conditions. The cellular functions that are disrupted by permutations in DISC1, which lead to the development of these disorders, have yet to be clearly defined and are the subject of current ongoing research.
Multiple DISC1 isoforms have been identified at the RNA level, including a TSNAX-DISC1 transgene splice variant, and at the protein level. Of the isolated RNA isomers, 4 have been confirmed to be translated namely Long form (L), Long variant isoform (Lv), Small isoform (S), and Especially small isoform (Es).
Human DISC1 is transcribed as two major splice variants, L form and Lv isoform. The L and Lv transcripts utilize distal and proximal splice sites, respectively, within exon 11. The L and Lv protein isoforms differ by only 22 amino acids within the C-terminus.
Alternate transcriptional splice variants, encoding different isoforms, have been characterized.
References:
1) Role of disrupted in schizophrenia 1 (DISC1) in stress-induced prefrontal cognitive dysfunction, 2013
2) Association analysis of the DISC1 gene with schizophrenia in the Japanese population and DISC1 immunoreactivity in the postmortem brain,2013
3) Revisiting disrupted-in-schizophrenia 1 as a scaffold protein,2013
4) Structure and evolutionary history of DISC1,2011
5) "Wikipedia 2015": http://en.wikipedia.org/wiki/DISC1
THE GENE

Wikigenes includes links to
- NCBI Gene
- NCBI SNP
- iHOP resource
- OMIM
- SNPedia
- UniProt
- Ensembl
- HGNC
The implication of genetics in psychiatric illnesses is not unique to schizophrenia, though the heritability of schizophrenia has been calculated as high as 80%.
The continued research of the family following the discovery of the translocation yielded statistical analysis of the probability of observing the simultaneous occurrence, or co-inheritance, of psychological afflictions and the translocation. This concept was measured quantitatively using the LOD, or logarithm of the odds value.
The higher the LOD value, the stronger the correlation between the presence of the translocation and given disease(s) is thought to be. The LOD for the chromosome 1 translocation and identification of schizophrenia alone in the Scottish family was found to be 3.6
The LOD value of the translocation and a broader number of diagnoses (including schizophrenia, schizoaffective disorder, bipolar affective disorder, and recurrent major depression) was found to be 7.1.
In a transcriptional profiling and genome-wide association analysis in 1232 pedigreed Mexican-American individuals for whom there were neuroanatomic images, neurocognitive assessments and neuropsychiatric diagnoses, SOLAR was used to determine heritability, identify gene expression patterns and perform association analyses on 188 quantitative brain-related phenotypes. The researchers found that the DISC1 transcript is highly heritable (h(2)=0.50; P=1.97 × 10(-22)), and that gene expression is strongly cis-regulated (cis-LOD=3.89) but is also influenced by trans-effects. They identified several DISC1 polymorphisms that were associated with cortical gray matter thickness within the parietal, temporal and frontal lobes. Associated regions affiliated with memory included the entorhinal cortex (rs821639, P=4.11 × 10(-5); rs2356606, P=4.71 × 10(-4)), cingulate cortex (rs16856322, P=2.88 × 10(-4)) and parahippocampal gyrus (rs821639, P=4.95 × 10(-4)); those affiliated with executive and other cognitive processing included the transverse temporal gyrus (rs9661837, P=5.21 × 10(-4); rs17773946, P=6.23 × 10(-4)), anterior cingulate cortex (rs2487453, P=4.79 × 10(-4); rs3738401, P=5.43 × 10(-4)) and medial orbitofrontal cortex (rs9661837; P=7.40 × 10(-4)). Cognitive measures of working memory (rs2793094, P=3.38 × 10(-4)), as well as lifetime history of depression (rs4658966, P=4.33 × 10(-4); rs12137417, P=4.93 × 10(-4)) and panic (rs12137417, P=7.41 × 10(-4)) were associated with DISC1 sequence variation. DISC1 has well-defined genetic regulation and clearly influences important phenotypes related to psychiatric disease
Besides large familial-based studies in which the pedigrees of various family members are examined, twin studies have also been a source of support for researchers in the investigation of DISC1.
In a meta-analysis of twin studies, twelve out of fourteen were found to support the fact that from a genetic prospective, schizophrenia is a complex trait that depends on both genetic and environmental factors.
Such findings have encouraged researchers to continue with both macro-analysis of the disorders afflicting individuals with the mutation, as well as explore the micro-level.
| Start: | 231,762,561 bp from pter |
| End: | 232,177,018 bp from pter |
| Size: | 414,458 bases |
| Orientation: | plus strand |
The DISC1 gene is situated at chromosome 1q42.1 and overlaps with DISC2 open reading frame.
DISC1 homologues have been identified in all major vertebrate families including the common chimpanzee, the Rhesus monkey, the house mouse, the brown rat, zebrafish,pufferfish, cattle, and dogs; additionally homologue's have been described for invertebrate and plant phyla.

References:
1) Impact of DISC1 variation on neuroanatomical and neurocognitive phenotypes,2011
2) OMIM
CHEMICAL STRUCTURE AND IMAGES
The protein encoded by this gene, is predicted to contain a coiled coil motif rich C-terminal domain and a N-terminal globular domain.
The N-terminus contains two putative nuclear localization signals and a serine-phenylalanine-rich motif of unknown significance.
The C-terminus contains multiple regions with coiled-coil forming potential and twoleucine zippers that may mediate protein-protein interactions.
Protein Aminoacids Percentage

SYNTHESIS AND TURNOVER
The protein locates to the nucleus, centrosome, cytoplasm, mitochondria, axons and synapses. Mitochondria are the predominant site of endogenous DISC1 expression, with at least two isoforms occupying internal mitochondrial locations. No known functional homologues exist for this protein in humans, although it does have broad homology to scaffold proteins. The DISC1 protein function appears to be highly diverse and its functional role in cellular processes is dependent upon the cellular domain it is located in. The presence or absence of certain protein interaction domains or targeting motifs may confer specific functions and influence sub cellular targeting, therefore it is probable that alternative splicing codetermines both the function and the intracellular location of DISC1
CELLULAR FUNCTIONS

Many studies have provided insight into the normal function of the DISC1 protein, though much remains to be clearly defined.
DISC1 is functionally involved in several processes that regulate neural development and brain maturation such as neuronal proliferation, differentiation, migration, cAMP signaling, cytoskeletal modulation, and translational regulation via various signaling pathways.
Much of what is understood about the normal function of DISC1 has been uncovered through studies on zebrafish and mice asmodel organisms.
In zebrafish, DISC1 is essential for forebrain development and GSK3/β-catenin signaling, while in mice the DISC1-GSK3 pathway regulates proliferation of neural progenitor cells in the cortex and adult dentate gyrus. This data suggests a direct DISC1 GSK3/β-catenin interaction.
Researches demonstrate using mouse, zebrafish, and human model systems that DISC1 variants are loss of function in Wnt/GSK3β signaling and disrupt brain development.

The DISC1 variants A83V, R264Q, and L607F, but not S704C, do not activate Wnt signaling compared with wild-type DISC1 resulting in decreased neural progenitor proliferation. In zebrafish, R264Q and L607F could not rescue DISC1 knockdown-mediated aberrant brain development. Furthermore, human lymphoblast cell lines endogenously expressing R264Q displayed impaired Wnt signaling.
Interestingly, S704C inhibited the migration of neurons in the developing neocortex. Data demonstrate DISC1 variants impair Wnt signaling and brain development and elucidate a possible mechanism for their role in neuropsychiatric phenotypes.
DISC1 functions through a rich protein-protein interaction network, named the “DISC1 interactome” by researchers.
Among its known interaction partners are 14-3-3ε, LIS1 and the PDE4B enzyme.
DISC1 may play an important role in neuroplasticity via interactions with molecules of the cytoskeleton and centrosome, such as NUDEL and LIS1. The protein also enables the activity of dynein, a microtubule protein. Controlling transport of microtubules is involved in neuronal migration, neurite outgrowth, and axon formation.

DISC1 is highly expressed during critical periods of brain development, particularly in the embryonic ventricular and subventricular zones of the cortex, where neural progenitor cells are found. This localization suggests that DISC1 is an important regulator of embryonic and adult neurogenesis, and may regulate proliferation and/or differentiation. Levels of the protein in cycling neural progenitor cells affects whether they differentiate into neurons or remain as progenitors. Expression profile is highest in the hippocampus during development and remains highly expressed in the adult dentate gyrus and olfactory bulb, regions where adult neurogenesis is present.
Reaserches found in zebrafish loss of DISC1 resulted in persistent CNC cell medial migration, dorsal to the developing neural epithelium, and hindered migration away from the region dorsal to the neural rod. General CNC cell motility was not affected by DISC1 knockdown, however, as the speed of CNC cells was indistinguishable from that of wild-type counterparts. They determined that the failure of CNC cells to migrate away from the neural rod correlated with the enhanced expression of two transcription factors, foxd3 and sox10. These transcription factors have many functions in CNC cells, including the maintenance of precursor pools, timing of migration onset, and the induction of cell differentiation. This work, in conjunction with previous studies, suggests that the perpetuation of expression of these factors affects several aspects of CNC cell development, leading to a loss of craniofacial cartilage and an expansion of peripheral cranial glia.
DISC1 has also been shown to regulate tempo of neuronal integration into the brain and guidance of positioning of new neurons.
Due to localization of the protein found at the synapse, DISC1 is also likely to play a key role in postsynaptic density, however this novel role is not yet fully understood.

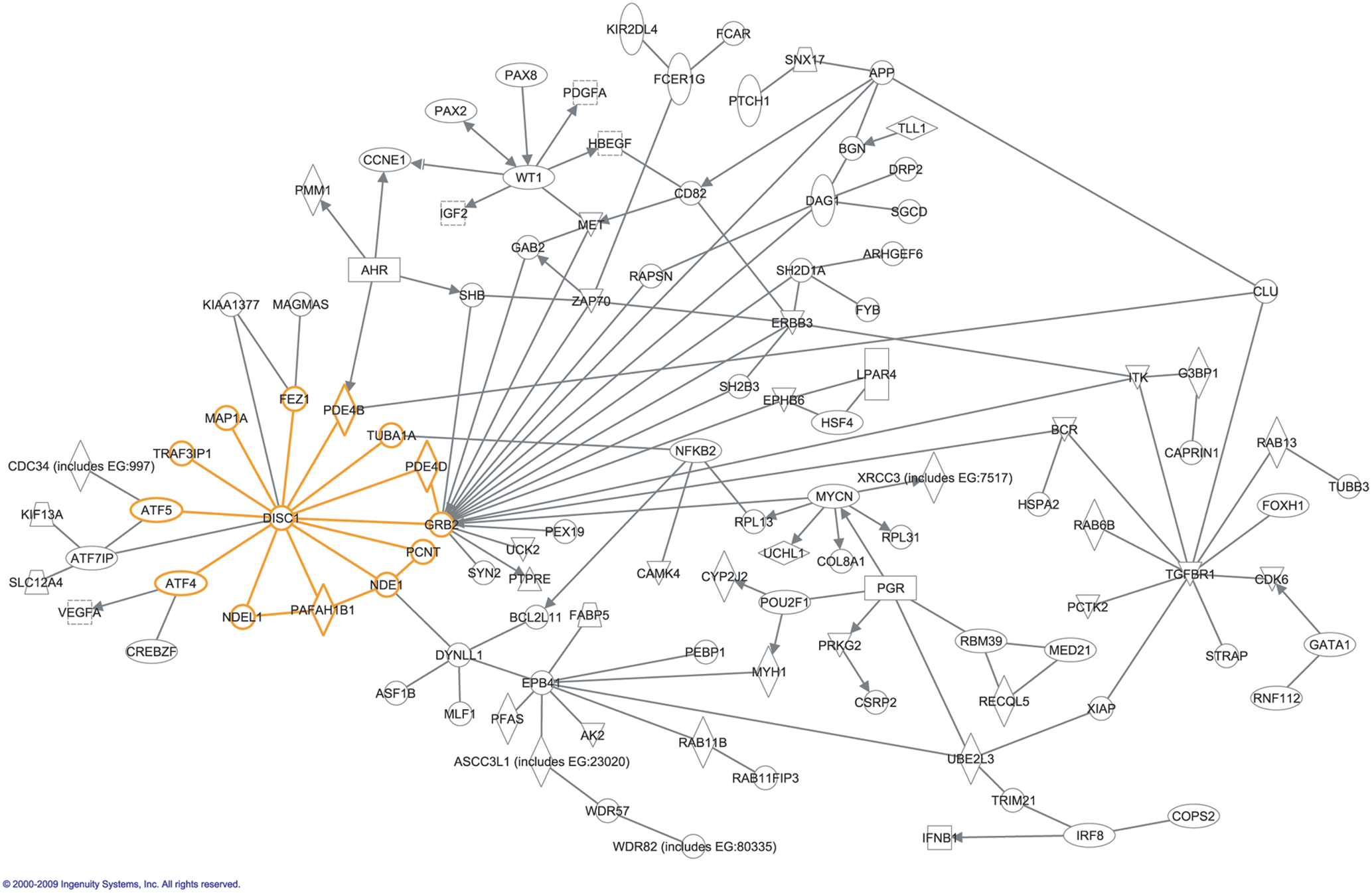
The DISC1 protein has no known enzymatic activity; rather it exerts its effect on multiple proteins through interactions to modulate their functional states and biological activities in time and space.
These include:
DISC1
DISC1 has been shown to self-associate, to form dimers, multimers, and oligomers. The ability of DISC1 to form complexes with itself may be important in regulating its affinity for interacting partners such as NDEL1.
In postmortem brain samples of Schizophrenia patients there is an increase in insoluble DISC1 oligomer aggregates, indicative of a common link with other neurological disorders characterised by protein aggregation, namely Alzheimer's Disease, Parkinson's Disease, and Huntington's Disease.
ATF4/ATF5
ATF4 and ATF5 are members of the leucine zipper activating transcription factor / CREB family. They are known to bind to and regulate the function of GABA receptors in synapses and are involved in signal transduction from the cell membrane to the nucleus. Both proteins interact with DISC1 and GABA receptors via their second C-terminal leucine zipper domain, thereforeDISC1 is able to regulate GABAB receptor function through its interaction with ATF4/ATF5.
FEZ1
DISC1 participates in neurite outgrowth through its interaction with the fasciculation and elongation protein ζ-1 (FEZ1).
FEZ1 is a mammalian homolog of the C. elegans UNC-76 protein involved in axonal outgrowth and fasciculation.
The C-terminal region of FEZ1 (aa 247-392) is required for interaction with DISC1. A DISC1 region (aa 446-633), containing two stretches with coiled-coil-forming potential is critical for its interaction with FEZ1.
DISC1-FEZ1 interaction is enhanced during neuro-differentiation, and expression of the FEZ1-binding domain of DISC1 has a dominant negative effect on neurite outgrowth, which implies co-operation of DISC1 and FEZ1 in this process.
Kalirin-7
The DISC1 protein plays a role in the process of regulating spine form and function through its interactions with kalirin-7 (kal-7).
Kal-7 is a regulator of spine morphology and synaptic plasticity in association with neuronal activity. Kal-7-dependent regulation of spine formation occurs through its activity as a GDP/GTP exchange factor for Rac1. Activation of rac1 by kal-7 leads to increased spine size and synaptic strength through regulation of the actin cytoskeleton by rac1.
DISC1 is able to bind to kal-7, confining its access to rac1, and in turn regulate spinal formation. Activation of NMDA receptors causes dissociation of DISC1 and kal-7, leaving kal-7 available to activate rac1.
MAP1A
DISC1 shows strong interaction with the microtubule-associated protein MAP1A that controls the polymerization and stabilization of microtubule networks in neurons, and therepy influence cell shape and intracellular transport of vesicles and organelles. MAP1A binds to the far N-terminus (aa 293-696) of DISC1, and the amino terminus of DISC1 binds to the LC2 subunit of MAP1A. The LC2 subunit of MAP1A contains an actin-binding domain and is necessary and sufficient for microtubule binding and polymerization, therefore DISC1 is able to regulate the ability of MAP1A to polymerize and stabilize microtubules and traffic proteins to their correct localization in the synaptic architecture.
NDEL1/NUDEL
DISC1 is localized to the centrosome, the primary microtubule organizing center of the cell, via interaction with nuclear distribution gene homologue-like 1 (NDEL1, also called NUDEL), where it is part of a protein complex involved in cytoskeletal processes of neuronal migration, including nucleokinesis and neurite outgrowth.
NUDEL is also known to play a role in axon regeneration and has an additional DISC1-modulated function as a cysteine endopeptidase.
Localization of NUDEL to axons is dependent on expression of DISC1.
NUDEL binds to a 100 amino acid domain of DISC1 (aa 598-697) containing a coiled coil domain and a leucine zipper. The amino acid domain of NUDEL that binds DISC1 is the carboxyl terminal 100 amino acids of the protein (aa 241-345), which contains a cytoplasmic dynein binding site.
PCM1/Pericentriolar Material
The protein Pericentriolar Material 1 (PCM1) which is associated with cilia development in the CNS interacts directly with the Disrupted-In-Schizophrenia 1 (DISC1) and calmodulin 1 (CALM1)proteins. Kamiya et al. have shown that PCM1, DISC1 and BBS4 can all disrupt neuronal organisation in the mouse when their expression is down-regulated. Markers at the pericentriolar material 1 gene (PCM1) have shown genetic association with schizophrenia in several schizophrenia case control studies. Resequencing of the genomic DNA from research volunteers who had inherited haplotypes associated with schizophrenia showed a threonine an isoleucine mis-sense mutation in exon 24 which may change the structure and function of PCM1 (rs370429).
This mutation was found only as a heterozygote in ninety eight schizophrenic research subjects and controls out of a total sample of 2,246 case and control research subjects. Amongst the ninety eight carriers of rs370429 sixty seven were affected with schizophrenia. The same alleles and haplotypes were associated with schizophrenia in both London and Aberdeen samples. Another potential aetiological base pair change in PCM1 was rs445422 which altered a splice site signal. A further mutation, rs208747, was shown by electrophoretic mobility shift assays to create or destroy a promoter transcription factor site. Five further non-synonymous changes in exons were also found. Given the number and identity of the haplotypes associated with schizophrenia further aetiological base pair changes must exist within and around the PCM1 gene. The findings in relation to PCM1 support the role of DISC1 also being a susceptibility locus for schizophrenia.
References:
1) Disc1 regulates foxd3 and sox10 expression, affecting neural crest migration and differentiation,2009
2) Common DISC1 polymorphisms disrupt Wnt/GSK3β signaling and brain development,2011
DIAGNOSTIC USE AND CLINICAL IMPLICATIONS
Aberrations of DISC1 are considered a generalized risk factor in major psychiatric diseases and have also been implicated in memory deficits and abnormal patterns of brain activity.
DISC1 translocation increases the risk of developing schizophrenia, bipolar disorder, or major depression by about 50-fold in comparison to the general population. Efforts to model DISC1 disease biology in transgenic mice, Drosophila, and zebrafish have provided psychiatric disease implications related to DISC1 mutations. However, no specific variant is consistently associated with development of mental disorders, indicating allelic heterogeneity in psychiatric disease. The impact of variants in the DISC1 gene on expression and protein function is not yet clearly defined and associated variants are not necessarily causative.
Schizophrenia affects 1% of the general population and is highly heritable, providing an indication of a genetic basis. DISC1 has been associated with neurological abnormalities such as delusions, deficits in long term and working memory, diminution of gray matter volume in hippocampal and prefrontal regions. These abnormalities are also seen as symptoms of schizophrenia. As DISC1 function is involved in neurogenesis and neuroplasticity, vulnerability to schizophrenia may involve dysfunction in the hippocampus, a brain region in which adult neurogenesis occurs.
In 2008, a genetic screen of 97 Finnish families affected by autism and Asperger’s syndrome revealed repeated DNA sequences within the DISC1 gene in those diagnosed with autism.
Furthermore, a single nucleotide change in the gene was found to be present in 83% of family members with Asperger’s syndrome. A recent family study has reported a large chromosome 1 deletion that includes loss of DISC1 in a young boy diagnosed with autism. A link between DISC1 duplication and autism has also been suggested by the finding of a seven-gene duplication that includes DISC1 carried by two brothers with autism and mild retardation. These alterations in people with the disorder are rare, however, as none were found in a screening of 260 Belgians with autism.
Transgenic model organism strains generated with mutated or absent DISC1 suggest that the gene may contribute to at least some autistic abnormalities. Mice with lowered levels of DISC1 expression exhibit abnormal response to electrical stimulation, a decrease of dopamine synthesis, and an inability to filter unnecessary sensory information. Studies of expression of mutant DISC1 prenatally and postnatally have demonstrated varying effects, indicating the possibility that early postnatal expression of mutant DISC1 causes features of autism. Many more studies are necessary to confirm these suggestion.
Linkage studies in extended families multiply affected with bipolar disorder also provide evidence for DISC1 as a genetic factor in the etiology of bipolar disorder. In 1998, a follow-up study was conducted of the large Scottish family in which DISC1 was first discovered. Additional family membe