21 05 20 fase2 il punto dell'epidemiologo Giuseppe Costa
.
30 04 2020 Documento CTS
.
15 04 20 Come un modello computazionale può aiutarci a gestire meglio l'epidemia da Covid-19
dove sono i risultati e la spiegazione del modello?
.
Mappa COVID-Piemonte
.
Modello Netlogo Terna et al.
.
Terna P., Pescarmona G., Acquadro A., Pescarmona P., Russo G., Terna S. (2020),
An Agent-Based Model of the Diffusion of Covid-19 Using NetLogo, URL
.
Home page Twitter
Dati Biologici Utili
COVID-19 DataPortal
.
Nasce il Covid-19 Open Research Dataset: 29mila articoli scientifici a disposizione dell’Ai 17 03 20
COVID-19 Italia
Coronavirus Knowledge Hub: A trusted source for the latest science on SARS-CoV-2 and COVID-19
OSCOVIDA: Open Science COVID Analysis Grafici casi, mortalità eccetera
pandemic-crisis-covid-19-fight-support-open-science-julien-hering
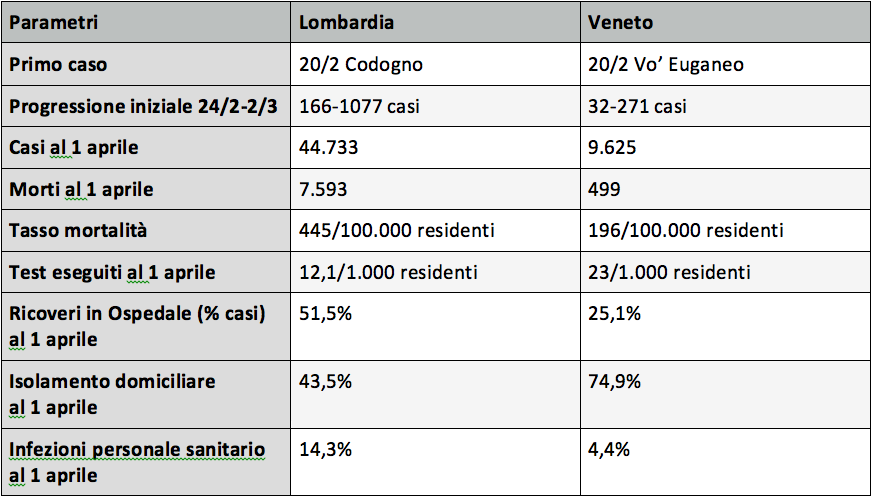
Tratto da: lombardia-e-veneto-due-approcci-confronto
Tratto da: inquinamento-e-covid-due-vaghi-indizi-non-fanno-prova
In seconda battuta, per infettare non bastano uno o pochi virus… ma deve esserci una definita carica virale sotto la quale non vi è infezione, come ribadito in modo assai convincente su Nature dal virologo della Charitè di Berlino, Christian Drosten [11].
Virological assessment of hospitalized patients with COVID-2019, 2020
The clinical courses in subjects under study were mild, all being youngto middle-aged professionals without significant underlying disease.
Apart from one patient, all cases were first tested when symptoms were
still mild or in the prodromal stage, a period in which most patients
would present once there is general awareness of a circulating pandemic disease5
. Diagnostic testing suggests that simple throat swabs
will provide sufficient sensitivity at this stage of infection. This is in
stark contrast to SARS. For instance, only 38 of 98 nasal or nasopharyngeal swab samples tested positive by RT-PCR in SARS patients in Hong
Kong15. Also, viral load differed considerably. In SARS, it took 7 to 10
days after onset until peak RNA concentrations (of up to 5×105
copies
per swab) were reached13,14. In the present study, peak concentrations
were reached before day 5, and were more than 1000 times higher.
Successful live virus isolation from throat swabs is another striking
difference from SARS, for which such isolation was rarely successful16–18.
Altogether, this suggests active virus replication in upper respiratory
tract tissues, where SARS-CoV is not thought to replicate in spite of
detectable ACE-2 expression19,20 . At the same time, the concurrent use
of ACE-2 as a receptor by SARS-CoV and SARS-CoV-2 corresponds to
a highly similar excretion kinetic in sputum, with active replication in
the lung. SARS-CoV was found in sputum at mean concentrations of 1.2-
2.8×106
copies per mL, which corresponds to observations made here13.
Whereas proof of replication by histopathology is awaited, extended
tissue tropism of SARS-CoV-2 with replication in the throat is strongly
supported by our studies of sgRNA-transcribing cells in throat swab
A C C E L E R A T E D A R TI C L E P R E VI E W
A C C E L E R A T E D A R TI C L E P R E VI E W
Nature | www.nature.com | 3
samples, particularly during the first 5 days of symptoms. Striking
additional evidence for independent replication in the throat is provided by sequence findings in one patient who consistently showed
a distinct virus in her throat as opposed to the lung. In addition, the
disturbance of gustatory and olfactory sense points at upper respiratory tract tissue infection.
Critically, the majority of patients in the present study seemed to be
already beyond their shedding peak in upper respiratory tract samples
when first tested, while shedding of infectious virus in sputum continued through the first week of symptoms. Together, these findings
suggest a more efficient transmission of SARS-CoV-2 than SARS-CoV
through active pharyngeal viral shedding at a time when symptoms
are still mild and typical of upper respiratory tract infection. Later in
the disease, COVID-19 then resembles SARS in terms of replication
in the lower respiratory tract. Of note, the two patients who showed
some symptoms of lung affection showed a prolonged viral load in
sputum. Our study is limited in that no severe cases were observed.
Future studies including severe cases should look at the prognostic
value of an increase of viral load beyond the end of week 1, potentially
indicating aggravation of symptoms.
One of the most interesting hypotheses to explain a potential extension of tropism to the throat is the presence of a polybasic furin-type
cleavage site at the S1-S2 junction in the SARS-CoV-2 spike protein that
is not present in SARS-CoV17. Insertion of a polybasic cleavage site in
the S1-S2 region in SARS-CoV was shown to lead to a moderate but
discernible gain of fusion activity that might result in increased viral
entry in tissues with low density of ACE2 expression21.
The combination of very high virus RNA concentrations and occasional detection of sgRNA-containing cells in stool indicate active replication in the gastrointestinal tract. Active replication is also suggested
by a much higher detection rate as compared to MERS-coronavirus,
for which we found stool-associated RNA in only 14.6% samples in 37
patients hospitalized in Riyadh, Saudi Arabia22,23. If virus was only passively present in stool, such as after swallowing respiratory secretions,
similar detection rates as for MERS-CoV would be expected. Replication
in the gastrointestinal tract is also supported by analogy with SARSCoV, which was regularly excreted in stool, from which it could be isolated in cell culture24. Our failure to isolate live SARS-CoV-2 from stool
may be due to the mild courses of cases, with only one case showing
intermittent diarrhea. In China, diarrhea has been seen in only 2 of 99
cases25. Further studies should therefore address whether SARS-CoV-2
shed in stool is rendered non-infectious though contact with the gut
environment. Our initial results suggest that measures to contain viral
spread should aim at droplet-, rather than fomite-based transmission.
The prolonged viral shedding in sputum is relevant not only for hospital infection control, but also for discharge management. In a situation
characterized by limited capacity of hospital beds in infectious diseases
wards, there is pressure for early discharge following treatment. Based
on the present findings, early discharge with ensuing home isolation
could be chosen for patients who are beyond day 10 of symptoms with
less than 100,000 viral RNA copies per ml of sputum. Both criteria predict that there is little residual risk of infectivity, based on cell culture.
The serological courses of all patients suggest a timing of seroconversion similar to or slightly earlier than in SARS-CoV infection18. Seroconversion in most cases of SARS occurred during the second week of
symptoms. As in SARS and MERS, IgM was not detected significantly
earlier than IgG in immunofluorescence, which might in part be due to
technical reasons as the higher avidity of IgG antibodies outcompetes
IgM for viral epitopes in the assay. IgG depletion can only partially
alleviate this effect. Because IFA is a labor-intensive method, ELISA
tests should be developed as a screening test. Neutralization testing
is necessary to rule out cross-reactive antibodies directed against
endemic human coronaviruses. Based on frequently low neutralizing
antibody titers observed in coronavirus infection26,27, we have here
developed a particularly sensitive plaque reduction neutralization
assay. Considering the titers observed, a simpler microneutralization
test format is likely to provide sufficient sensitivity in routine application and population studies.
When aligned to viral load courses, it seems there is no abrupt virus
elimination at the time of seroconversion. Rather, seroconversion early
in week 2 coincides with a slow but steady decline of sputum viral load.
Whether certain properties such as glycosylation pattern at critical
sites of the glycoprotein play a role in the attenuation of neutralizing
antibody response needs further clarification. In any case, vaccine
approaches targeting mainly the induction of antibody responses
should aim to induce particularly strong antibody responses in order
to be effective.
Online content
Dilution Effect