DEFINITION
Angiotensin-converting enzyme 2 / ACE2 is an enzyme attached to the outer surface (cell membranes) of cells in the lungs, arteries, heart, kidney, and intestines. ACE2 lowers blood pressure by catalyzing the cleavage of angiotensin II (a vasoconstrictor peptide) into angiotensin (1–7) (a vasodilator). ACE2 also serves as the entry point into cells for some coronaviruses.
THE GENE
x+chromosome+inactivation+ace+2
ACE2 is located on X Chromosome at Xp22, location were most genes escape silencing.
Atlas of Genetics and Cytogenetics in Oncology and Haematology: ACE2
Genes that escape from X inactivation, 2011
To achieve a balanced gene expression dosage between males (XY) and females (XX), mammals have evolved a compensatory mechanism to randomly inactivate one of the female X chromosomes. Despite this chromosome-wide silencing, a number of genes escape X inactivation: in women about 15% of X-linked genes are bi-allelically expressed and in mice, about 3%. Expression from the inactive X allele varies from a few percent of that from the active allele to near equal expression. While most genes have a stable inactivation pattern, a subset of genes exhibit tissue-specific differences in escape from X inactivation. Escape genes appear to be protected from the repressive chromatin modifications associated with X inactivation. Differences in the identity and distribution of escape genes between species and tissues suggest a role for these genes in the evolution of sex differences in specific phenotypes. The higher expression of escape genes in females than in males implies that they may have female-specific roles and may be responsible for some of the phenotypes observed in X aneuploidy.
About 10% of X-linked genes but only 3% of auto-somal genes cause intellectual disability when mutated (Ropers 2008; Ropers and Hamel 2005; Zechner et al. 2001)
Human genes escaping X-inactivation revealed by single cell expression data, 2019
Table Table comparing annotation of all 75 ChrX escape candidates.
Genes That Escape X-Inactivation in Humans Have High Intraspecific Variability in Expression, Are Associated with Mental Impairment but Are Not Slow Evolving, 2013
An additional Table listing genes that escape inactivation
X-Chromosome Inactivation: A Crossroads Between Chromosome Architecture and Gene Regulation 2018
In somatic nuclei of female therian mammals, the two X chromosomes display very different chromatin states: One X is typically euchromatic and transcriptionally active, and the other is mostly silent and forms a cytologically detectable heterochromatic structure termed the Barr body. These differences, which arise during female development as a result of X-chromosome inactivation (XCI), have been the focus of research for many decades. Initial approaches to define the structure of the inactive X chromosome (Xi) and its relationship to gene expression mainly involved microscopy-based approaches. More recently, with the advent of genomic techniques such as chromosome conformation capture, molecular details of the structure and expression of the Xi have been revealed. Here, we review our current knowledge of the 3D organization of the mammalian X-chromosome chromatin and discuss its relationship with gene activity in light of the initiation, spreading, and maintenance of XCI, as well as escape from gene silencing.
CHEMICAL STRUCTURE AND IMAGES
When relevant for the function
- Primary structure
- Secondary structure
- Tertiary structure
- Quaternary structure
Protein Aminoacids Percentage (Width 700 px)


SYNTHESIS AND TURNOVER
mRNA synthesis
protein synthesis
post-translational modifications
degradation
CELLULAR FUNCTIONS
cellular localization,
biological function
- Cell signaling and Ligand transport
- Structural proteins
REGULATION
DIAGNOSTIC USE
AMP-Activated Protein Kinase Regulates Endothelial Cell Angiotensin-Converting Enzyme Expression via p53 and the Post-Transcriptional Regulation of microRNA-143/145, 2013
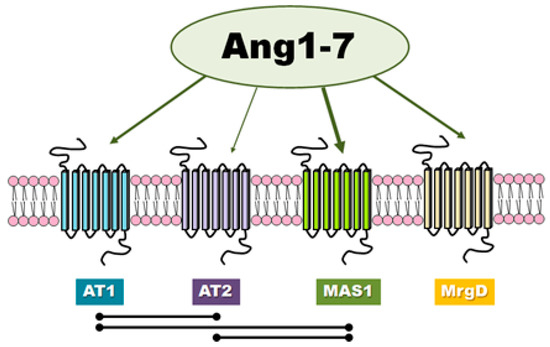
Involvement of ACE2/Ang-(1-7)/MAS1 Axis in the Regulation of Ovarian Function in Mammals, 2020
MAS1 has a Glu/Gln ratio of 3, pointing to dependence on estrogens.
More MAS1 women have healthier
Aerobic training-mediated DNA hypermethylation of Agtr1a and Mas1 genes ameliorate mesenteric arterial function in spontaneously hypertensive rats, 2021
Protein Aminoacids Percentage (Width 700 px)



ACE2/Ang-(1-7)/Mas1 axis and the vascular system: Vasoprotection to COVID-19-associated vascular disease, 2021