Kegg Pathways
Aspartic acid is in equilibrium with glutamic acid via AST

Aspartic acid is in equilibrium with Asparagine via Asparagine Synthetase and Asparaginase
Asparagine Function
Asparagine promotes cancer cell proliferation through use as an amino acid exchange factor, 2016.
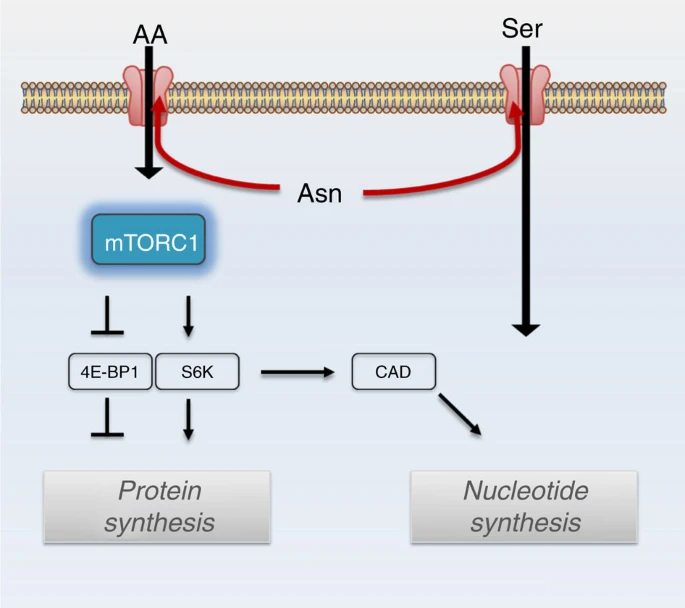
- Abstract
Cellular amino acid uptake is critical for mTOR complex 1 (mTORC1) activation and cell proliferation. However, the regulation of amino acid uptake is not well-understood. Here we describe a role for asparagine as an amino acid exchange factor: intracellular asparagine exchanges with extracellular amino acids. Through asparagine synthetase knockdown and altering of media asparagine concentrations, we show that intracellular asparagine levels regulate uptake of amino acids, especially serine, arginine and histidine. Through its exchange factor role, asparagine regulates mTORC1 activity and protein synthesis. In addition, we show that asparagine regulation of serine uptake influences serine metabolism and nucleotide synthesis, suggesting that asparagine is involved in coordinating protein and nucleotide synthesis. Finally, we show that maintenance of intracellular asparagine levels is critical for cancer cell growth. Collectively, our results indicate that asparagine is an important regulator of cancer cell amino acid homeostasis, anabolic metabolism and proliferation.
Asparagine Synthetase and Asparaginase regulate the ratio aspartate/asparagine
Asparagine Synthetase
It is an irreversible reaction

asparagine synthetase and induction
Metabolic Alterations Caused by KRAS Mutations in Colorectal Cancer Contribute to Cell Adaptation to Glutamine Depletion by Upregulation of Asparagine Synthetase. 2016
Activation of the amino acid response modulates lineage specification during differentiation of murine embryonic stem cells. 2013
AAR activation caused the expected increase in transcription factors that mediate specific AAR pathways, as well as the induction of asparagine synthetase, a terminal AAR target gene.
Using several human CRC cell lines and clinical specimens of primary CRC, we demonstrated that the expression of asparagine synthetase (ASNS), an enzyme that synthesizes asparagine from aspartate, was upregulated by mutated KRAS and that ASNS expression was induced by KRAS-activated signaling pathway, in particular PI3K-AKT-mTOR pathway.
Asparaginase

THE GENES
THE GENE
CHEMICAL STRUCTURE AND IMAGES
When relevant for the function
- Primary structure
- Secondary structure
- Tertiary structure
- Quaternary structure
h3. SYNTHESIS AND TURNOVER
mRNA synthesis
protein synthesis
post-translational modifications
degradation
CELLULAR FUNCTIONS
cellular localization,
biological function
- Cell signaling and Ligand transport
- Structural proteins
REGULATION
DIAGNOSTIC USE


Asparagine Synthetase
ASNS_HUMAN
Brenda[]=Homo+sapiens&show_tm=0

Amino Acid Deprivation and Endoplasmic Reticulum Stress Induce Expression of Multiple Activating Transcription Factor-3 mRNA Species That, When Overexpressed in HepG2 Cells, Modulate Transcription by the Human Asparagine Synthetase Promoter 2002
Transcription from the ASNS (asparagine synthetase) gene is increased in response to either amino acid (amino acid response) or glucose (endoplasmic reticulum stress response) deprivation. These two independent pathways converge on the same set of genomic cis-elements within the ASNS promoter, referred to as nutrient-sensing response element-1 and -2.
Enhanced expression of asparagine synthetase under glucose-deprived conditions protects pancreatic cancer cells from apoptosis induced by glucose deprivation and cisplatin. 2007
Asparaginase (exogenous addiction)
Asparaginase is an enzyme that catalyzes the hydrolysis of asparagine to aspartic acid, which undergoes transamination with alfa-ketoglutarate to yield glutamate and oxaloacetate.

Enhanced expression of asparagine synthetase under glucose-deprived conditions protects pancreatic cancer cells from apoptosis induced by glucose deprivation and cisplatin. 2007
Transcriptional regulation of the human asparagine synthetase gene by carbohydrate availability. 1999
Transcription of the asparagine synthetase (AS) gene is induced by amino acid deprivation.
Regulation of asparagine synthetase gene transcription by the basic region leucine zipper transcription factors ATF5 and CHOP. 2005
Here we show that overexpression of the bZIP protein ATF5, a transcriptional activator, stimulates asparagine synthetase promoter/reporter gene transcription via the nutrient-sensing response unit.
Induction of asparagine synthetase by follicle-stimulating hormone in primary cultures of rat Sertoli cells. 1994
The results suggest that asparagine synthetase is induced via a cAMP-dependent signal transduction pathway in Sertoli cells.
Induction of asparagine synthetase during lymphocyte activation by phytohemagglutinin. 1989
The increase of asparagine synthetase activity was inhibited by cycloheximide and somewhat by actinomycin D, suggesting de novo enzyme synthesis during the stimulation.
GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent L-asparaginase. 2009
Asparaginase depletes circulating asparagine and glutamine, activating amino acid deprivation responses (AADR) such as phosphorylation of eukaryotic initiation factor 2 (p-eIF2) leading to increased mRNA levels of asparagine synthetase and CCAAT/enhancer-binding protein β homologous protein (CHOP) and decreased mammalian target of rapamycin complex 1 (mTORC1) signaling.
Asparagine synthetase as a causal, predictive biomarker for L-asparaginase activity in ovarian cancer cells. 2006
L-Asparaginase (l-ASP), a bacterial enzyme used since the 1970s to treat acute lymphoblastic leukemia, selectively starves cells that cannot synthesize sufficient asparagine for their own needs. Molecular profiling of the NCI-60 cancer cell lines using five different microarray platforms showed strong negative correlations of asparagine synthetase (ASNS) expression and DNA copy number with sensitivity to l-ASP in the leukemia and ovarian cancer cell subsets. To assess whether the ovarian relationship is causal, we used RNA interference to silence ASNS in three ovarian lines and observed 4- to 5-fold potentiation of sensitivity to l-ASP with two of the lines. For OVCAR-8, the line that expresses the least ASNS, the potentiation was >500-fold. Significantly, that potentiation was >700-fold in the multidrug-resistant derivative OVCAR-8/ADR, showing that the causal relationship between ASNS expression and l-ASP activity survives development of classical multidrug resistance. Tissue microarrays confirmed low ASNS expression in a subset of clinical ovarian cancers as well as other tumor types. Overall, this pharmacogenomic/pharmacoproteomic study suggests the use of l-ASP for treatment of a subset of ovarian cancers (and perhaps other tumor types), with ASNS as a biomarker for patient selection.
Fun about asparaginase
ASGL1_HUMAN
FUNCTION: Has both L-asparaginase and beta-aspartyl peptidase activity. May be involved in the production of L-aspartate, which can act as an excitatory neurotransmitter in some brain regions. Is highly active with L-Asp beta-methyl ester. Besides, has catalytic activity toward beta-aspartyl dipeptides and their methyl esters, including beta-L-Asp-L-Phe, beta-L-Asp-L-Phe methyl ester (aspartame), beta-L-Asp-L-Ala, beta-L-Asp-L-Leu and beta-L- Asp-L-Lys. Does not have aspartylglucosaminidase activity and is inactive toward GlcNAc-L-Asn. Likewise, has no activity toward glutamine.
CATALYTIC ACTIVITY: L-asparagine + H(2)O = L-aspartate + NH.
CATALYTIC ACTIVITY: Cleavage of a beta-linked Asp residue from the N-terminus of a polypeptide.
ENZYME REGULATION: Glycine accelerates autocleavage into an alpha and beta chain.
BIOPHYSICOCHEMICAL PROPERTIES: Kinetic parameters: KM=3.4 mM for L-asparagine (L-Asn); KM=0.4 mM for L-aspartic acid beta-methyl ester; KM=0.4 mM for L-Asp-L-Phe; KM=1.0 mM for L-Asp-L-Ala;
SUBUNIT: Heterodimer of an alpha and beta chain produced by autocleavage. This heterodimer may then dimerize in turn, giving rise to a heterotetramer.
SUBCELLULAR LOCATION: Cytoplasm. Note=Midpiece of sperm tail.
ALTERNATIVE PRODUCTS: Event=Alternative splicing; Named isoforms=2; Name=1; IsoId=Q7L266-1; Sequence=Displayed; Name=2; IsoId=Q7L266-2; Sequence=VSP_028287; Note=No experimental confirmation available;
TISSUE SPECIFICITY: Expressed in brain, kidney, testis and tissues of the gastrointestinal tract. Present in sperm (at protein level). Over-expressed in uterine, mammary, prostatic and ovarian carcinoma.
INDUCTION: By 5-alpha-di-hydrotestosterone and progesterone.
PTM: Cleaved into an alpha and beta chain by autocatalysis; this activates the enzyme. The N-terminal residue of the beta subunit is responsible for the nucleophile hydrolase activity.
SIMILARITY: Belongs to the Ntn-hydrolase family.
SEQUENCE CAUTION: Sequence=BAB15302.1; Type=Erroneous initiation; Note=Translation N-terminally extended; Sequence=BAB15302.1; Type=Miscellaneous discrepancy; Note=Contaminating sequenc