DEFINITION
HMG-CoA reductase (or 3-hydroxy-3-methyl-glutaryl-CoA reductase or HMGCR) is the rate-controlling enzyme (EC 1.1.1.88) of the mevalonate pathway
THE GENE
CHEMICAL STRUCTURE AND IMAGES
When relevant for the function
- Primary structure
- Secondary structure
- Tertiary structure
- Quaternary structure

HMG-CoA reductase is a resident glycoprotein of the endoplasmic reticulum (ER) that consists of two distinct domains:
- a hydrophobic N-terminal domain with eight membrane-spanning regions
- a hydrophilic C-terminal domain that projects into the cytosol.
The N-terminal domain of HMG-CoA reductase, which anchors the protein in membrane of the ER, contains sites for Insig binding and sterol-regulated ubiquitination (lysines 89 and 248). The region is both necessary and sufficient for sterol-accelerated degradation.
The C-terminal domain of HMG-CoA reductase exerts all of the enzymatic activity and has no role in regulated degradation of the enzyme.
Protein Aminoacids Percentage
The Protein Aminoacids Percentage gives useful information on the local environment and the metabolic status of the cell (starvation, lack of essential AA, hypoxia)
Protein Aminoacids Percentage (Width 700 px)

SYNTHESIS AND TURNOVER
mRNA synthesis
protein synthesis
post-translational modifications
degradation
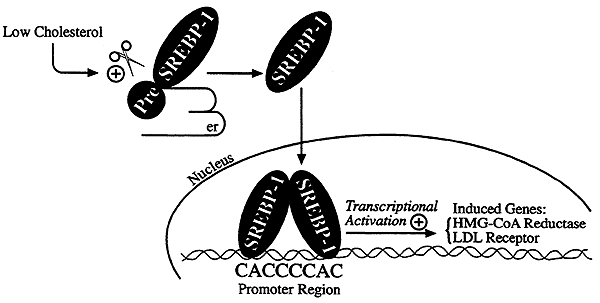
Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase 2010
Multiple mechanisms for feedback control of cholesterol synthesis converge on the rate-limiting enzyme in the pathway, 3-hydroxy-3-methylglutaryl coenzyme A reductase. This complex feedback regulatory system is mediated by sterol and nonsterol metabolites of mevalonate, the immediate product of reductase activity. One mechanism for feedback control of reductase involves rapid degradation of the enzyme from membranes of the endoplasmic reticulum (ER). This degradation results from the accumulation of sterols in ER membranes, which triggers binding of reductase to ER membrane proteins called Insig-1 and Insig-2. Insig binding leads to the recruitment of a membrane-associated ubiquitin ligase called gp78 that initiates ubiquitination of reductase. Ubiquitinated reductase then becomes extracted from ER membranes and is delivered to cytosolic 26S proteasomes through an unknown mechanism that is mediated by the gp78-associated ATPase Valosin-containing protein/p97 and appears to be augmented by nonsterol isoprenoids. Here, we will highlight several advances that have led to the current view of mechanisms for sterol-accelerated, ER-associated degradation of reductase. In addition, we will discuss potential mechanisms for other aspects of the pathway such as selection of reductase for gp78-mediated ubiquitination, extraction of the ubiquitinated enzyme from ER membranes, and the contribution of Insig-mediated degradation to overall regulation of reductase in whole animals.
CELLULAR FUNCTIONS
cellular localization,
biological function
- Cell signaling and Ligand transport
- Structural proteins
REGULATION

Potential use of HMG-CoA reductase inhibitors (statins) as radioprotective agents, 2010

Lipid droplets: a unified view of a dynamic organelle 2006
HMG-CoA Reductase Inhibitors Suppress Intracellular Calcium Mobilization and Membrane Current Induced by Lysophosphatidylcholine in Endothelial Cells 2002
DIAGNOSTIC USE



Mechanism for oxygen sensing in the cholesterol synthetic pathway. The link between synthesis of cholesterol and oxygen sensing in animal cells is provided by hypoxia-induced accumulation of lanosterol and 24,25-dihydrolanosterol and HIF-1alpha-mediated induction of Insig-1 and Insig-2. Convergence of these responses leads to rapid degradation of HMG CoA reductase, thereby limiting synthesis of cholesterol.

Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase 2008
PDF