Structure


Coenzyme Q synthesis requires:
- Tyrosine + Vit B6, Ascorbic Acid, Pantothenic Acid
- Clinical implications of the correlation between coenzyme Q10 and vitamin B6 status, 2016
- Abstract. The endogenous biosynthesis of the quinone nucleus of coenzyme Q10 (CoQ10) from tyrosine is dependent on adequate vitamin B6 nutriture. Lowered blood and tissue levels of CoQ10 have been observed in a number of clinical conditions. Many of these clinical conditions are most prevalent among the elderly. Kalen et al. have shown that blood levels of CoQ10 decline with age. Similarly, Kant et al. have shown that indicators of vitamin B6 status also decline with age. Blood samples were collected from 29 patients who were not currently being supplemented with either CoQ10 or vitamin B6. Mean CoQ10 concentrations was 1.1 ± 0.3 µg/ml of blood. Mean specific activities of EGOT was 0.30 ± 0.13 µmol pyruvate/hr/108 erythrocytes and the mean percent saturation of EGOT with PLP was 78.2 ± 13.9%. Means for all parameters were within normal ranges. Strong positive correlation was found between CoQ10 and the specific activity of EGOT (r = 0.5787, p < 0.001) and between CoQ10 and the percent saturation of EGOT with PLP (r = 0.4174, p < 0.024). Studies are currently in progress to determine the effect of supplementation with vitamin B6 of blood CoQ10 levels. It appears prudent to recommend that patients receiving supplemental CoQ10 be concurrently supplemented with vitamin B6 to provide for better endogenous synthesis of CoQ10 along with the exogenous CoQ10.
- 1. Introduction
In 1983, Olson and Rudney reviewed the work which led to the elucidation of the biosynthetic pathway for coenzyme Q (Fig. 1). Most higher organisms have lost the ability to produce non-steroidal aromatic compounds from simple precursors. They, instead, rely on dietary precursors for aromatic compounds. Coenzyme Q (CoQ) biosynthesis begins with the conversion of tyrosine to 4-hydroxybenzoic acid. 4-Hydroxybenzoic acid is the key precursor to the benzoquinone nucleus of CoQ. The isoprenoid side chain is added at the next step and its length is variable according to species. Humans and most mammals synthesize CoQ10.
Vitamin B6, as pyridoxal 50-phosphate (PLP), is required for the initial transamination step which produces 4-hydroxyphenylpyruvic acid from tyrosine. Thus, an adequacy of vitamin B6 nutriture is essential for the synthesis of CoQ.
- Farnesyl diphosphate (via Polyprenyldiphosphate)
- S-adenosyl methionine


Kegg Pathway
CoQ10 = 10 isoprenoid units
1 isoprenoid units ( 2 NADPH + 3 ATP each )
- 2 AcetylCoA + 2 NADPH --> mevalonate
- mevalonate + 3 ATP --> isopentenyl pirophosphate + 3 ADP + 1 CO2
1 4-hydroxybenzoate
- K00500 phenylalanine-4-hydroxylase [EC:1.14.16.1] 1 NADPH + 1 O2
- K03181 chorismate--pyruvate lyase [EC:4.1.3.-] [RN:R01302]

- K06125 4-hydroxybenzoate hexaprenyltransferase [EC:2.5.1.-]
[RN:R05000] Coq2
- K00591 hexaprenyldihydroxybenzoate methyltransferase [EC:2.1.1.114]
[RN:R08771] Coq3 1 SAME
- K06126 ubiquinone biosynthesis monooxygenase Coq6 [EC:1.14.13.-]
[RN:R08773] 1 NADPH + 1 O2
- K06127 ubiquinone biosynthesis methyltransferase [EC:2.1.1.-]
[RN:R08774] 1 SAME
- K06134 ubiquinone biosynthesis monooxygenase Coq7 [EC:1.14.13.-]
[RN:R08775] 1 NADPH + 1 O2
- K00591 hexaprenyldihydroxybenzoate methyltransferase [EC:2.1.1.114]
[RN:R08781] 1 SAME
Total Balance for 1 CoQ10 molecules:
Cell localization
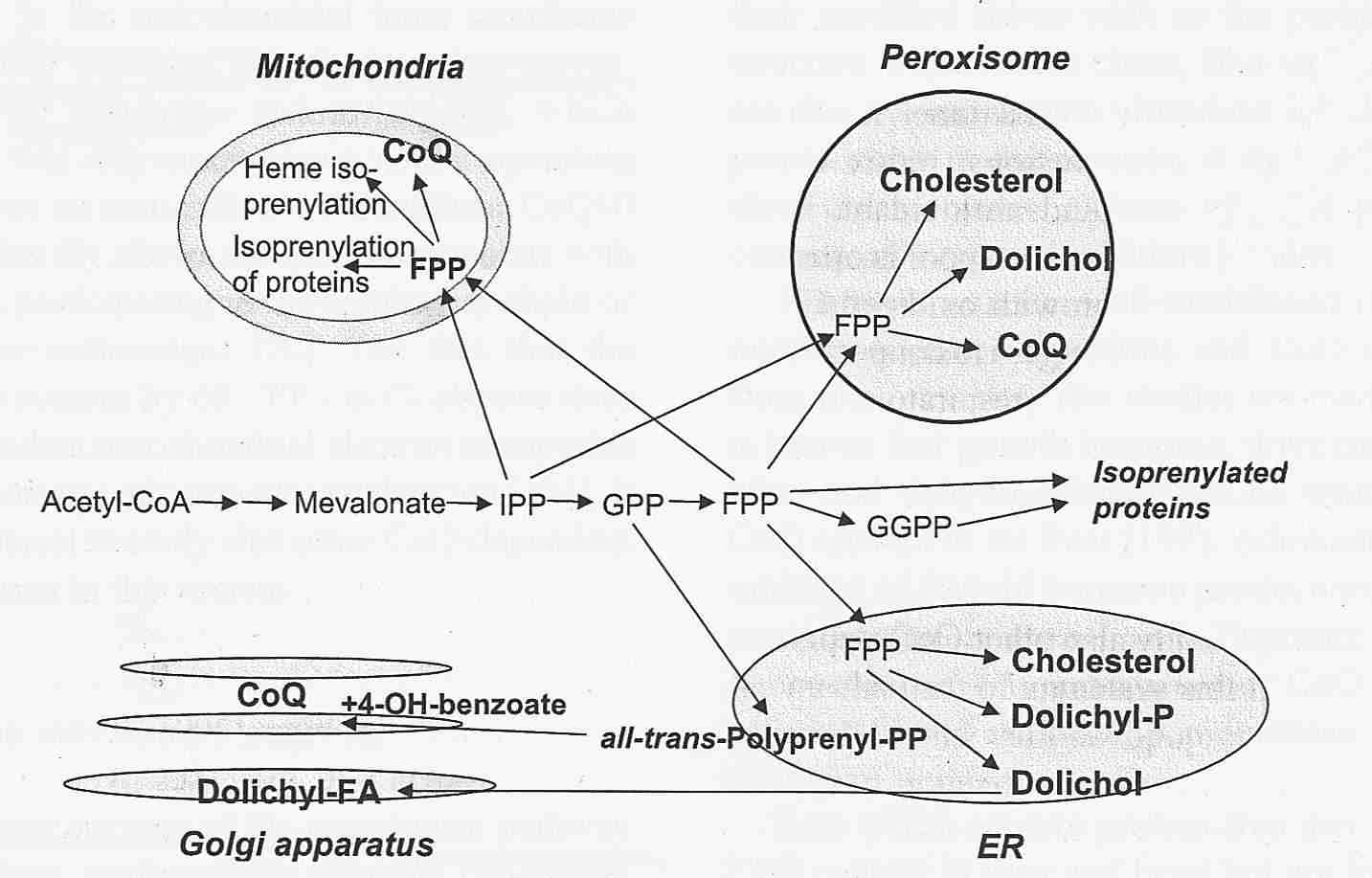
Endogenous Synthesis of Coenzyme Q in Eukaryotes 2007

Coq1 through Coq9 polypeptides localize to the mitochondria. In vitro mitochondria import were investigated for seven of the yeast Coq polypeptides and demonstrated to be dependent on a mitochondrial membrane potential (Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q synthesis. 2001: The putative mitochondrial-targeting sequence present at the amino-terminus of the polypeptide efficiently imported it to mitochondria in a membrane-potential-dependent manner). The function and submitochondrial localization of the nine Coq proteins, required for Q biosynthesis in eukaryotes is summarized in Table 1. A model incorporating genetic and physical evidence for a yeast Q biosynthetic multi-subunit complex is shown.
Local synthesis of nuclear-encoded mitochondrial proteins in the presynaptic nerve terminal. 2001
J Neurosci Res. 2001 Jun 1;64(5):447-53.
Gioio AE, Eyman M, Zhang H, Lavina ZS, Giuditta A, Kaplan BB.
One of the central tenets in neuroscience has been that the protein constituents of distal compartments of the neuron (e.g., the axon and nerve terminal) are synthesized in the nerve cell body and are subsequently transported to their ultimate sites of function. In contrast to this postulate, we have established previously that a heterogeneous population of mRNAs and biologically active polyribosomes exist in the giant axon and presynaptic nerve terminals of the photoreceptor neurons in squid. We report that these mRNA populations contain mRNAs for nuclear-encoded mitochondrial proteins to include: cytochrome oxidase subunit 17, propionyl-CoA carboxylase (EC 6.4.1.3), dihydrolipoamide dehydrogenase (EC 1.8.1.4), and coenzyme Q subunit 7. The mRNA for heat shock protein 70, a chaperone protein known to be involved in the import of proteins into mitochondria, has also been identified. Electrophoretic gel analysis of newly synthesized proteins in the synaptosomal fraction isolated from the squid optic lobe revealed that the large presynaptic terminals of the photoreceptor neuron contain a cytoplasmic protein synthetic system. Importantly, a significant amount of the cycloheximide resistant proteins locally synthesized in the terminal becomes associated with mitochondria. PCR analysis of RNA from synaptosomal polysomes establishes that COX17 and CoQ7 mRNAs are being actively translated. Taken together, these findings indicate that proteins required for the maintenance of mitochondrial function are synthesized locally in the presynaptic nerve terminal, and call attention to the intimacy of the relationship between the terminal and its energy generating system. J. Neurosci. Res. 64:447-453, 2001.
Axonal protein synthesis and the regulation of local mitochondrial function. 2009 Fulltext
Blood mononuclear cell coenzyme Q10 concentration and mitochondrial respiratory chain succinate cytochrome-c reductase activity in phenylketonuric patients. 2002 J Inherit Metab Dis. 2002 Dec;25(8):673-9.
- Coenzyme Q10 (CoQ10) serves as an electron carrier within the mitochondrial respiratory chain (MRC), where it is integrally involved in oxidative phosphorylation and consequently ATP production. It has recently been suggested that phenylketonuria (PKU) patients may be susceptible to a CoQ10 deficiency as a consequence of their phenylalanine-restricted diet, which avoids foods rich in CoQ10 and its precursors. Furthermore, the high phenylalanine level in PKU patients not on dietary restriction may also result in impaired endogenous CoQ10 production, as previous studies have suggested an inhibitory effect of phenylalanine on HMG-CoA reductase, the rate-controlling enzyme in CoQ10 biosynthesis. We investigated the effect of both dietary restriction and elevated plasma phenylalanine concentration on blood mononuclear cell CoQ10 concentration and the activity of MRC complex II + III (succinate:cytochrome-c reductase; an enzyme that relies on endogenous CoQ10) in a PKU patient population. The concentrations of CoQ10 and MRC complex II + III activity were not found to be significantly different between the PKU patients on dietary restriction, PKU patients off dietary restriction and the control group, although plasma phenylalanine levels were markedly different. The results from this investigation suggest that dietary restriction and the elevated plasma phenylalanine levels of PKU patients do not effect mononuclear cell CoQ10 concentration and consequently the activity of complex II + III of the MRC.
Regulation
Polyisoprenoid epoxides stimulate the biosynthesis of coenzyme Q and inhibit cholesterol synthesis. 2008
Drugs affecting CoQ10 synthesis
Amitriptyline induces coenzyme Q deficiency and oxidative damage in mouse lung and liver., 2011
Potentiation of the toxicity of adriamycin by propranolol. 1978
Both propranolol and adriamycin are biochemically known to inhibit mitochondrial CoQ10-enzymes of myocardial tissue in vitro. Both propranolol and adriamycin are clinically known to cause cardiotoxicity. At two dose levels of propranolol which caused no deaths to mice when administered alone, significant potentiation (p less than 0.01) of the lethality of adriamycin to mice was observed. These data, projected to the clinical situation, seem to contraindicate the administration of the beta-blocker, propranolol, for the hypertension of a cancer patient who is being treated with adriamycin.
Coenzyme Q10 and cardiovascular disease: a review. 2002
It appears that levels of coenzyme Q10 are decreased during therapy with HMG-CoA reductase inhibitors, gemfibrozil, Adriamycin, and certain beta blockers.
Mevalonic acid products as mediators of cell proliferation in simian virus 40-transformed 3T3 cells. 1987
- Effects of treatment with serum-free medium and 25-hydroxycholesterol (25-OH) on the cell cycle of simian virus 40-transformed 3T3 fibroblasts, designated SV-3T3 cells, were studied and compared with simultaneous effects on the activity of 3-hydroxy-3-methylglutaryl (HMG) CoA reductase and incorporation of [3H]mevalonic acid into cholesterol, Coenzyme Q, and dolichol. The data confirm our previous finding (O. Larsson and A. Zetterberg, Cancer Res., 46: 1233-1239, 1986) that 25-OH inhibits the cell cycle traverse of SV-3T3 cells specifically in early G1. In contrast, treatment with serum-free medium had no effect on cell cycle progression. The effect of 25-OH on the cell cycle traverse was correlated to a substantial decrease in the activity of HMG CoA reductase, whereas there was no change in the rate of [3H]mevalonic acid incorporated into cholesterol, Coenzyme Q, and dolichol. When the cells were exposed to serum-free medium, there was no depression of activity of HMG CoA reductase, and the rate of [3H]mevalonic acid incorporated into dolichol and cholesterol was not affected in any appreciable degree. In contrast the rate of Coenzyme Q synthesis was substantially decreased as a result of serum depletion (iron depletion?). A similar decrease in Coenzyme Q synthesis was also achieved by treating the cells with cholesterol-poor serum. This indicates that the rate of Coenzyme Q synthesis is dependent on the concentration of cholesterol in the culture medium. In order to analyze whether some of the products in the mevalonic acid biosynthetic pathway may be of importance in the control of G1 traverse and cell proliferation of SV-3T3 cells, cholesterol, Coenzyme Q, and dolichol were added as supplements to cells treated with 25-OH. It was shown that dolichol was capable of overcoming the 25-OH-induced inhibition of G1 traverse efficiently, whereas cholesterol and Coenzyme Q were considerably less effective. Considered together with the fact that the activity of HMG CoA reductase and incorporation of mevalonic acid into dolichol were unaffected following serum-free treatment, the results suggest that maintenance of a certain level of de novo synthesis of dolichol may contribute to the capability of SV-3T3 cells to proliferate in serum-free medium.